 Image credit: Unsplash
Image credit: UnsplashWhy We Need Better Health Text Datasets
The rise of the internet has revolutionized how we access health information. A staggering 89% of patients in the U.S. now google their health symptoms before visiting their doctor. This reliance on online health advice underscores the importance of accurately modeling and understanding this information. However, current health entity recognizers, tools that extract and classify health-related entities from text, fall short. They are primarily trained on clinical notes, which differ significantly from the language and content found in public health data. This means they often fail to accurately represent entities relevant to public health data, leaving a gap in our understanding and utilization of online health advice.
To address this, we introduce HealthE, a new annotated dataset designed specifically for the modeling of online health advice. HealthE consists of 3,400 health advice statements with token-level entity annotations, and an additional 2,256 health statements which are not health advice. The annotation scheme of HealthE is unique in its inclusion of entities important to modeling informal health texts where health concepts are mentioned in layman’s terms, such as food and exercise. This dataset is the first of its kind, designed to facilitate the large-scale analysis of online textual health advice.
The goal of our research is to improve the detection and classification of health advice and relevant entities in online health communities. This includes recognizing whether a text contains actionable health advice and identifying the entities in the text that pertain to relevant health concepts. By doing so, we aim to enhance health literacy, medical information search, and patient empowerment, and contribute to the development of more advanced patient-centric health applications.
How We Created HealthE
We began by scraping over 30,000 candidate advice statements from four reputable online health sources: WebMD, Medline Plus, CDC, and Covid Protocols. From these, a random sample of 4,500 candidates was selected for annotation by a team of three human annotators. The annotators classified each statement as either “Advice” or “Not Advice”, with 2,244 out of the 4,500 statements identified as “Advice” by at least two annotators. This process demonstrated substantial inter-annotator agreement, a key indicator of the reliability of the annotations. In addition to the newly collected data, we also incorporated 1,156 samples from the Preclude dataset, a public dataset of health advice statements.
The final HealthE dataset contains 3,400 health advice statements, each annotated for six entity types: Food/Nutrient, Disease/Condition, Medicine/Supplement/Treatment, Exercise, Vitals/Physiological Status, and Other. The entity annotation process was carried out by a team of 14 computational health researchers, ensuring the quality and accuracy of the dataset. Of the 13,989 labeled entity instances, 5,418 entities are unique, with the Medicine/Supplement/Treatment class having the highest number of unique entities.
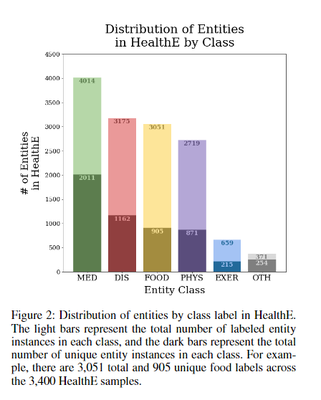
The HealthE dataset is a comprehensive resource for health entity recognition, designed specifically for online health communities and patient-centric information systems.
Benchmarking Results
To facilitate future research using HealthE, we benchmark both the Health Advice Classification (HAC) and Health Entity Recognition (HAC) tasks.
Health Advice Classification
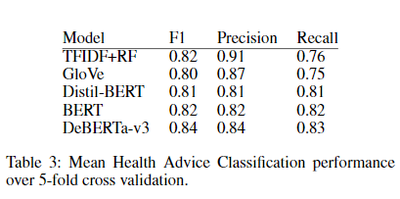
We evalaute classification by using two statistical text vectorization models and three transformer-based models. The transformer models, DistilBERT, BERT, and DeBERTa-v3, represent a diverse set of state-of-the-art models for large language model fine-tuning. Interestingly, all models performed well on HAC, with DeBERTa-v3 showing the best performance. However, even the lightweight models like TFIDF+RF achieved high F1 scores, indicating that simple statistical models can perform similarly to transformer models on HAC.
Health Entity Recognition
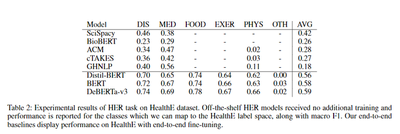
We also investigated the capacity of five popular HER models to classify the HealthE dataset. These models, including Amazon Comprehend Medical and Google Health NLP, are known for their effectiveness in extracting medical/health entity information from clinical notes and biomedical research publications. However, we found that these models often struggle with the HealthE label space, particularly with abstract terms and unfamiliar medical treatments.
On the contrary, our end-to-end (E2E) HER models performed reasonably well across all classes except ‘Other’. DeBERTa-v3 showed the best performance, with a macro F1 score of 0.59 across all classes. These findings highlight the usefulness of HER annotations in HealthE while showcasing the lack of domain transfer from biomedical entity recognition to HER.
Why it Matters
The ability to automatically identify and classify health entities in natural language has improved the performance of various systems, and HealthE is poised to further enhance this. The dataset’s representation of data from online health communities like WebMD and HealthLine is crucial for building patient-centric health management tools. Improved HER via HealthE can benefit works building patient-centric tools, as it provides a more comprehensive understanding of health advice in the online space.
HealthE is a significant contribution to the field of patient-centric health NLP, and we hope it will facilitate the detection of health advice statements and extraction of health entities in Health NLP applications.
Interested researchers are encouraged to download and explore HealthE to start building better Health NLP tools!